6 Notes
6.1 Stamp function
The stamp function can be used to highlight the nodes with colored rectangles.
library(ggkegg)
ggraph(pathway("ko00270"), layout="manual", x=x, y=y)+
geom_node_rect()+
stamp("ko:K00789")+
theme_minimal()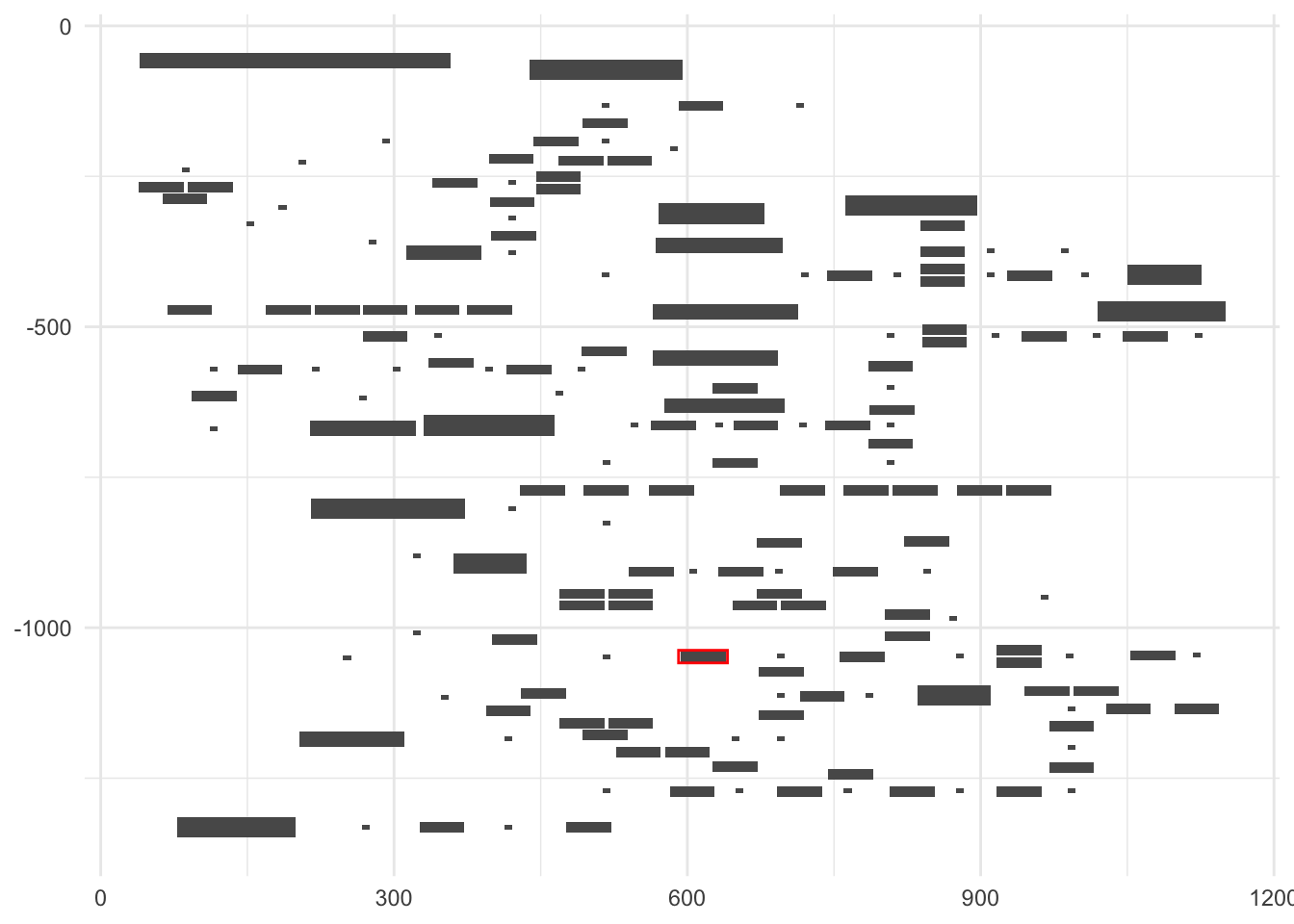
6.2 Parsing of the module definition
For module describing multiple lines of the definition (e.g. M00897), the module function tries to parse them separately. These definitions will be assigned the number (in character type).
mod <- module("M00899")
defs <- get_module_attribute(mod, "definitions")
defs[["1"]]
#> $definition_block
#> [1] "K00878"
#>
#> $definition_kos
#> [1] "K00878"
#>
#> $definition_num_in_block
#> [1] 1
#>
#> $definition_ko_in_block
#> $definition_ko_in_block[[1]]
#> [1] "K00878"
defs[["2"]]
#> $definition_block
#> [1] "((K00941 K00788),K14153,K21219)"
#>
#> $definition_kos
#> [1] "K00941" "K00788" "K14153" "K21219"
#>
#> $definition_num_in_block
#> [1] 4
#>
#> $definition_ko_in_block
#> $definition_ko_in_block[[1]]
#> [1] "K00941" "K00788" "K14153" "K21219"
## Extract definition 2
mod |>
module_text("2") |> ## return data.frame
plot_module_text() ## wrapper function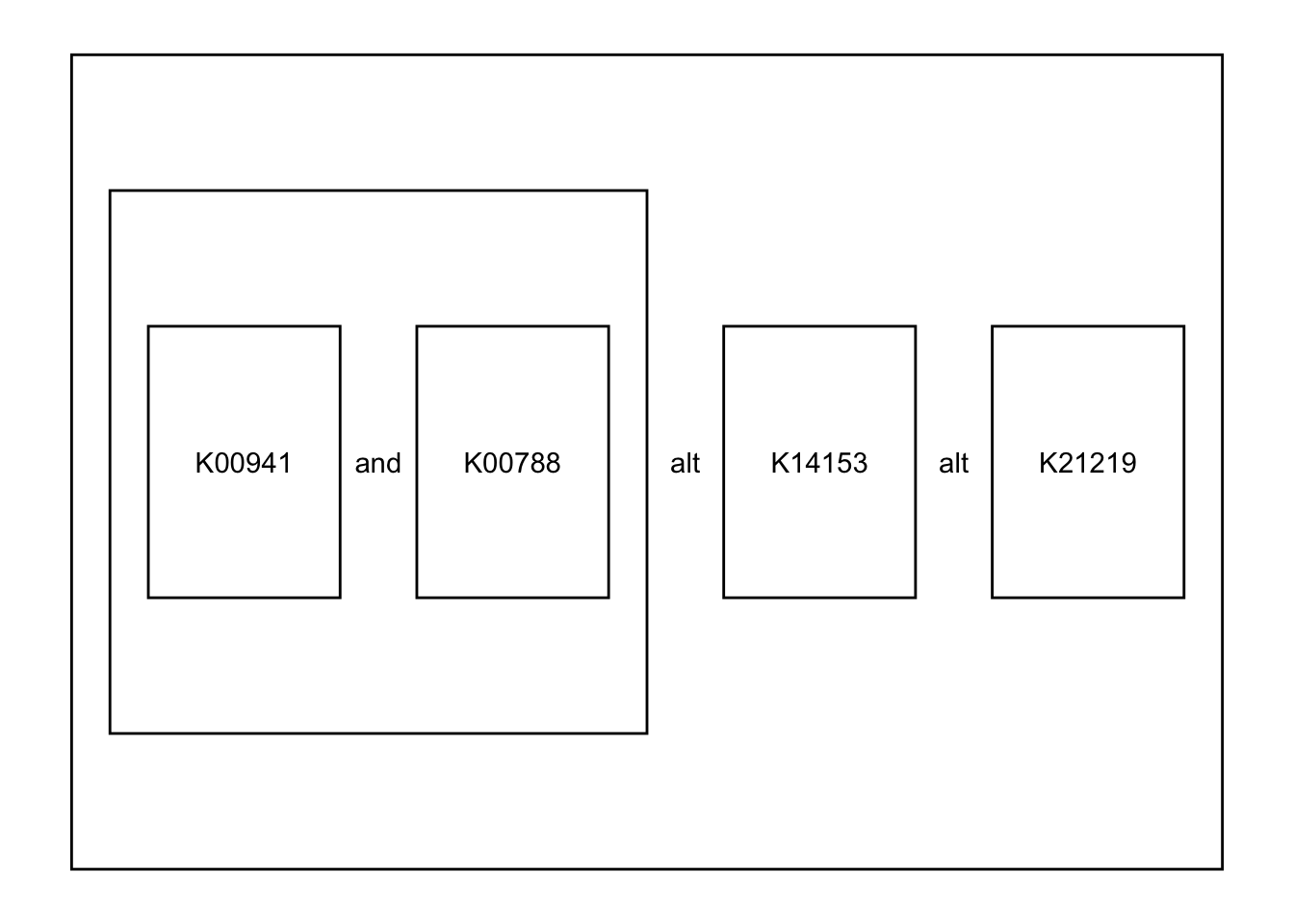
6.3 Parsing of reactions in the pathway
In the devel branch of ggkegg, the KGML of the following reactions (https://www.genome.jp/entry/R00863) in ko00270 are parsed in the below format by default.
<reaction id="237" name="rn:R00863" type="irreversible">
<substrate id="183" name="cpd:C00606"/>
<product id="238" name="cpd:C00041"/>
<product id="236" name="cpd:C09306"/>
</reaction>
library(ggkegg)
reac <- pathway("ko00270")
nname <- reac %N>% pull(name)
reac %E>% data.frame() %>% filter(reaction=="rn:R00863") %>%
mutate(fromn=nname[from], ton=nname[to])
#> from to type subtype_name subtype_value
#> 1 91 128 irreversible substrate <NA>
#> 2 128 129 irreversible product <NA>
#> 3 128 127 irreversible product <NA>
#> reaction reaction_id pathway_id fromn ton
#> 1 rn:R00863 237 ko00270 cpd:C00606 ko:K09758
#> 2 rn:R00863 237 ko00270 ko:K09758 cpd:C00041
#> 3 rn:R00863 237 ko00270 ko:K09758 cpd:C09306These edges correspond to the relationship between compound substrate or product and reaction (orthology) nodes. These edges are kept for the conversion in process_reaction, where substrate and product are directly connected by edges.
reac2 <- reac |> process_reaction()
node_df <- reac2 %N>% data.frame()
reac2 %E>% data.frame() |>
filter(reaction=="rn:R00863") |>
mutate(from_name=node_df$name[from], to_name=node_df$name[to])
#> from to type subtype_name subtype_value
#> 1 91 129 irreversible <NA> <NA>
#> 2 91 127 irreversible <NA> <NA>
#> reaction reaction_id pathway_id name bgcolor
#> 1 rn:R00863 <NA> <NA> ko:K09758 #BFBFFF
#> 2 rn:R00863 <NA> <NA> ko:K09758 #BFBFFF
#> fgcolor from_name to_name
#> 1 #000000 cpd:C00606 cpd:C00041
#> 2 #000000 cpd:C00606 cpd:C093066.4 Interpolation in the raster image
The annotation_custom is used in overlay_raw_map. The interpolate option can be specified.
ggraph(pathway("hsa04110"), layout="manual", x=x, y=y) + overlay_raw_map(interpolate=TRUE) + theme_void()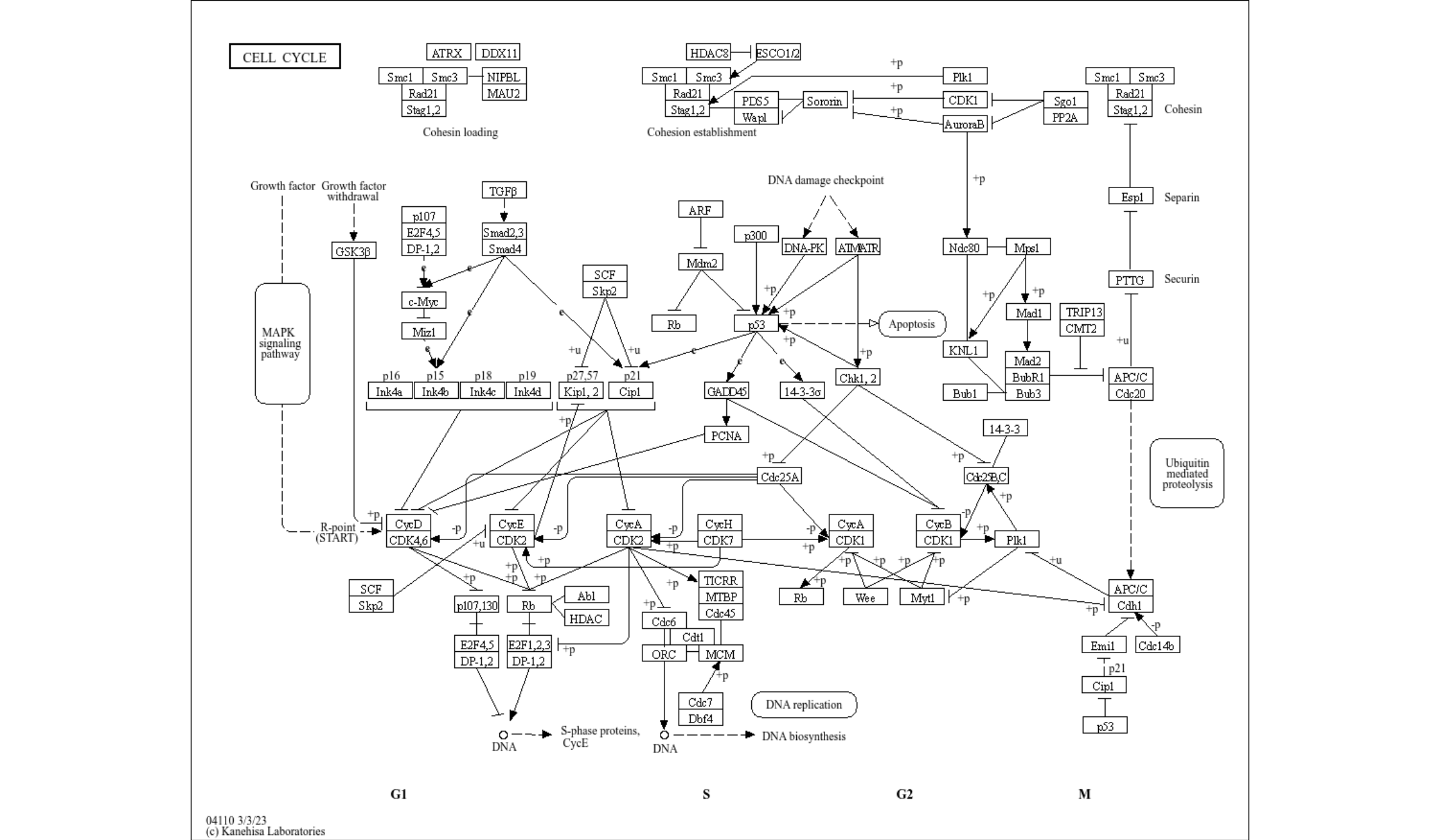
ggraph(pathway("hsa04110"), layout="manual", x=x, y=y) + overlay_raw_map(interpolate=FALSE) + theme_void()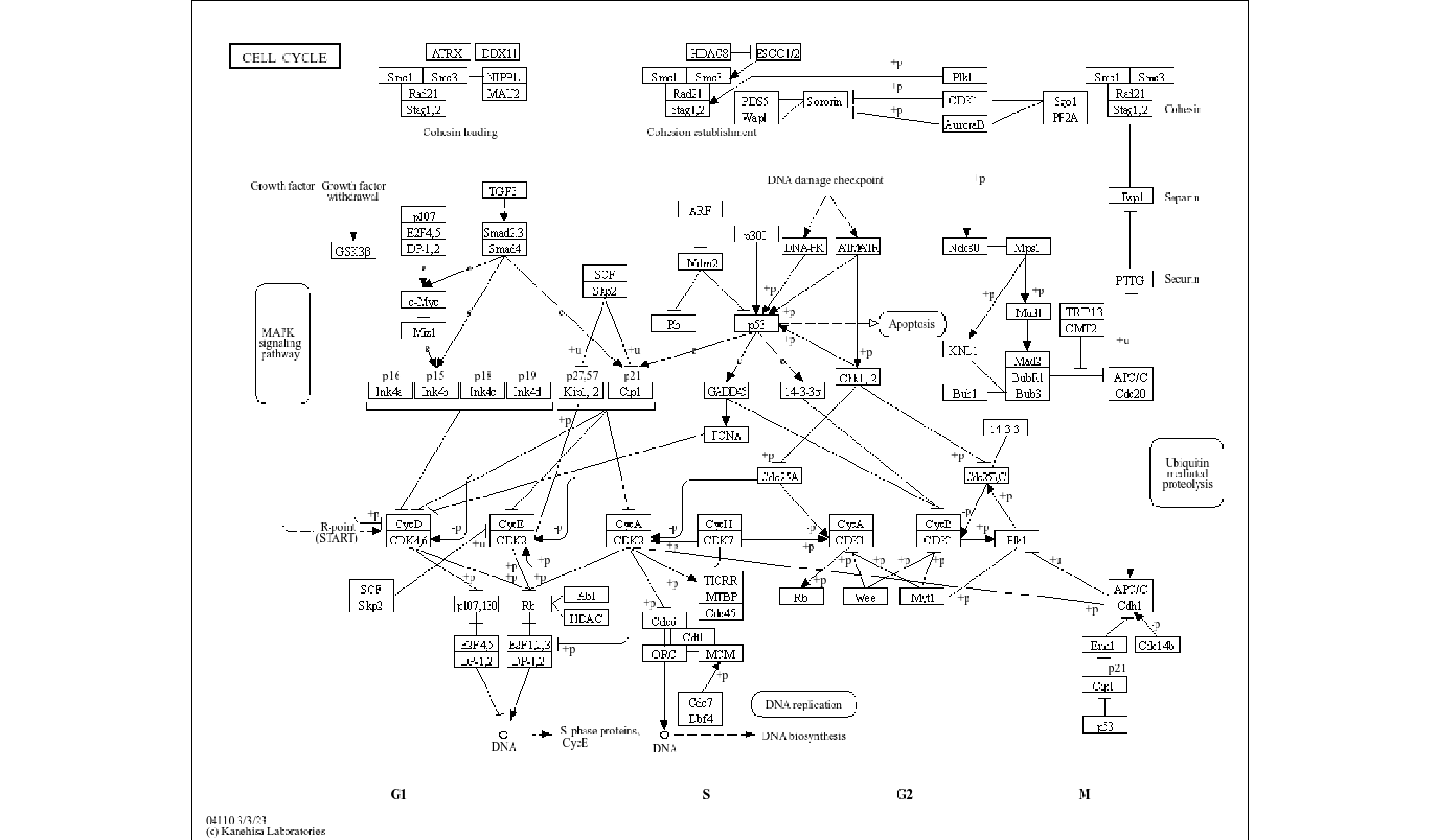
6.5 add_readable_edge_label
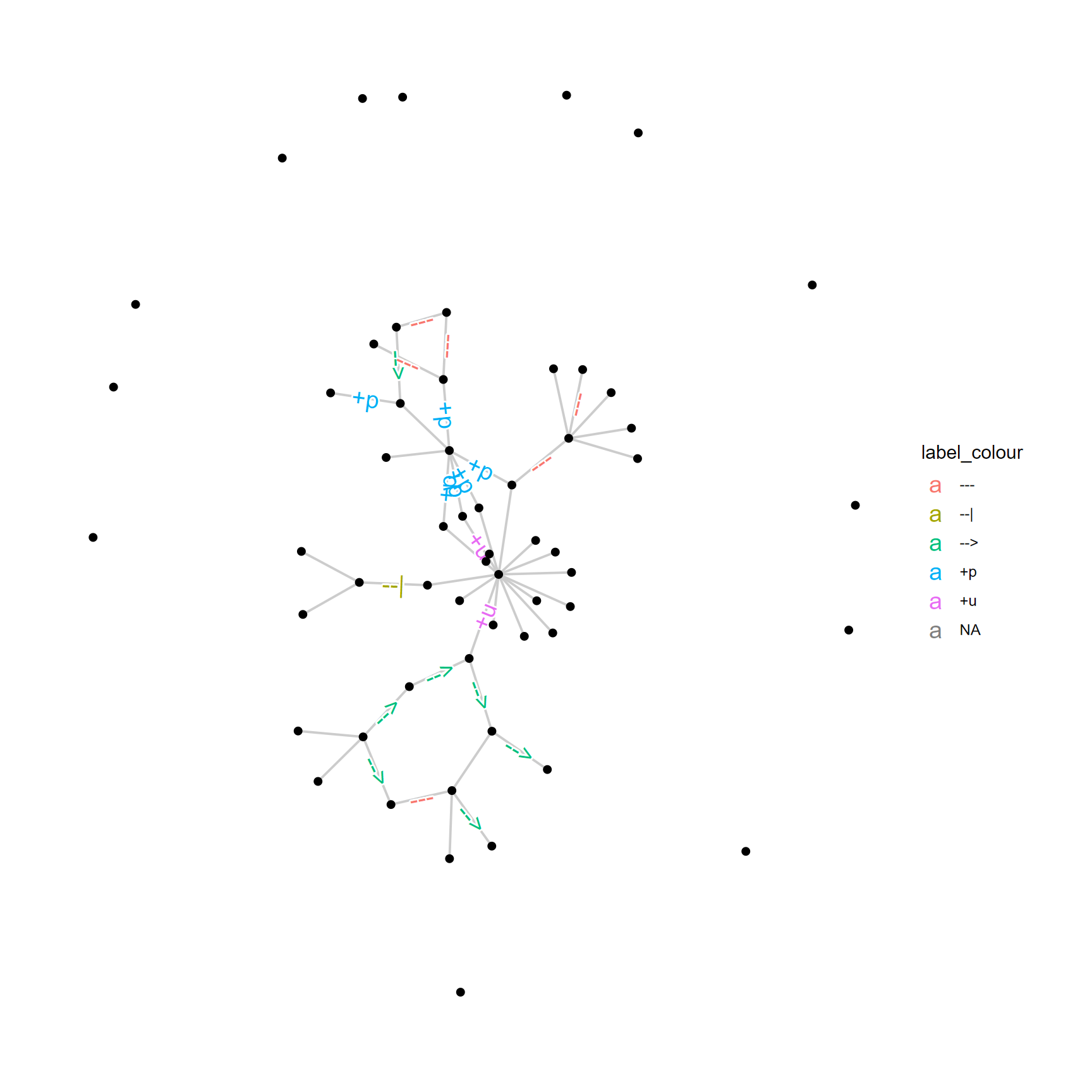
The example in 5.6 uses customized ggraph function for highlighting edge labels. add_readable_edge_label can highlight the edge label by shadowtext. This layer must be inserted just after the layer with labels. label_colour argument in the mapping should specify which column in the edge to be used for coloring.
pathway("hsa03460") %>%
ggraph(layout="kk")+
geom_edge_link(aes(label=subtype_value,
label_colour=subtype_value), color="grey80")+
add_readable_edge_label(size=5)+
geom_node_point()+
theme_graph()