1 About
ggkegg fetches information from KEGG and parse, analyze and visualize them using ggplot2 and ggraph, combined with the other packages investigating biological functions using KEGG. This package aims to visualize the complex components of KEGG using the grammar of graphics. For Python, please use pykegg using plotnine, which offers the almost same functionality as ggkegg used in conjuction with the package such as gseapy and PyDESeq2 and single-cell transcriptomics analysis library scanpy.
ggkegg uses KEGG API, and is restricted to academic use by academic users belonging to academic institutions.
One of the main aims of ggkegg is manupilating KEGG information in tidy ways using tidygraph, and offers the customized visualization of KEGG information including KEGG PATHWAY, MODULE, and NETWORK.
library(dplyr)
library(tidygraph)
pathway("hsa04110") |> ## Obtain and parse the KEGG pathway
activate(nodes) |> ## node manipulation
mutate(convert_hsa=convert_id("hsa"),
convert_map=convert_id("pathway")) |> ## convert IDs for organism hsa and pathway
ggraph(x=x, y=y)+ ## ggraph plot
geom_edge_parallel(arrow=arrow(length=unit(1,"mm")),
aes(linetype=subtype_name),
end_cap=circle(7.5,"mm"))+ ## Parallel edges
geom_node_rect(aes(filter=type=="gene",
fill=I(bgcolor)),
color="black")+ ## rectangular nodes
geom_node_text(aes(label=convert_hsa),
size=2, family="serif")+ ## text
geom_node_text(aes(label=convert_hsa,
filter=!is.na(convert_hsa) & convert_hsa=="TP53"),
size=2, color="red", family="serif")+ ## highlight text
theme_void()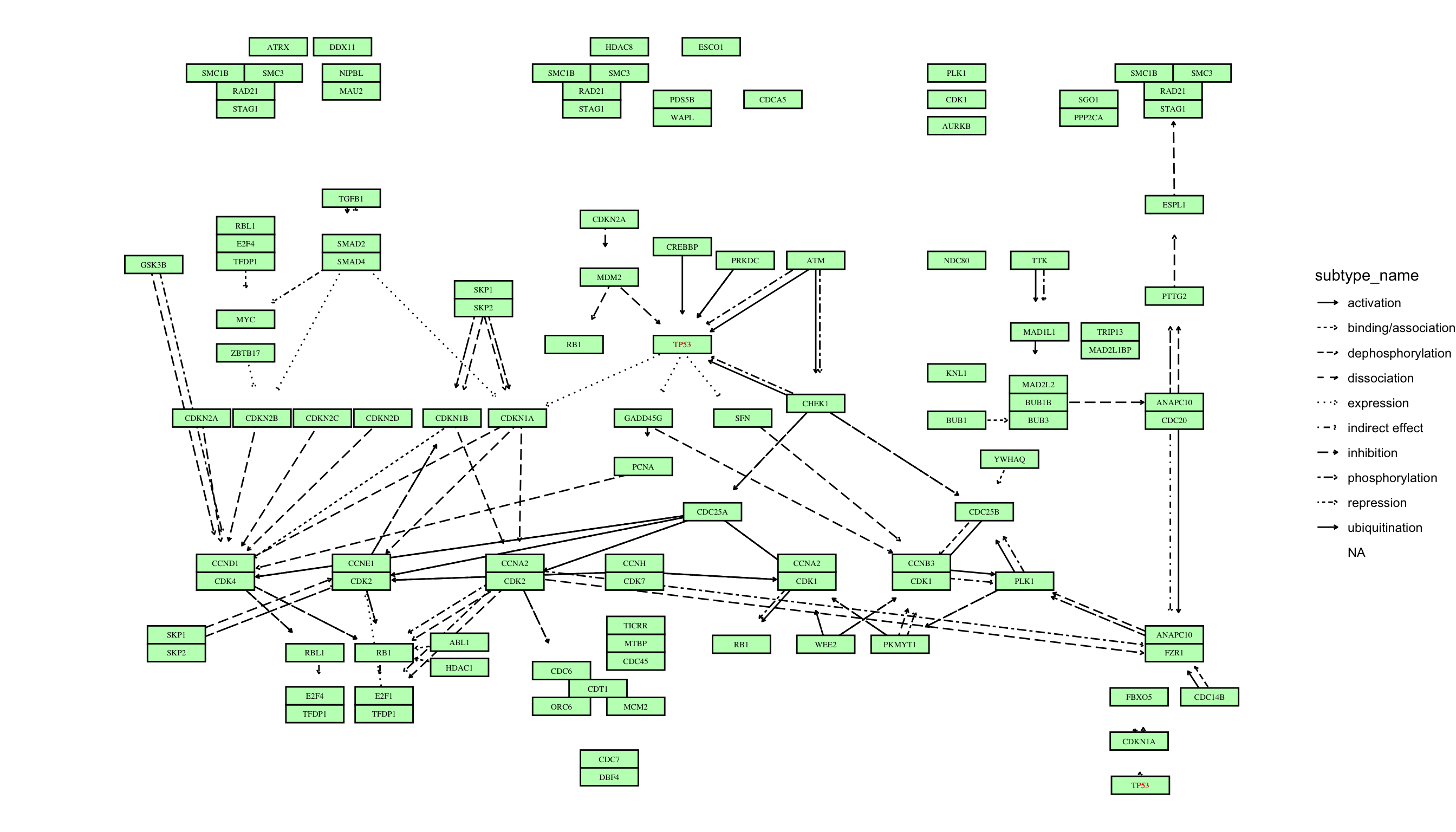
Some convenient functions are prepared, like highlighting genes in the pathway.
## Highlight genes in the pathway, and overlay raw map.
highlight_entities("hsa04110", "CDKN2A")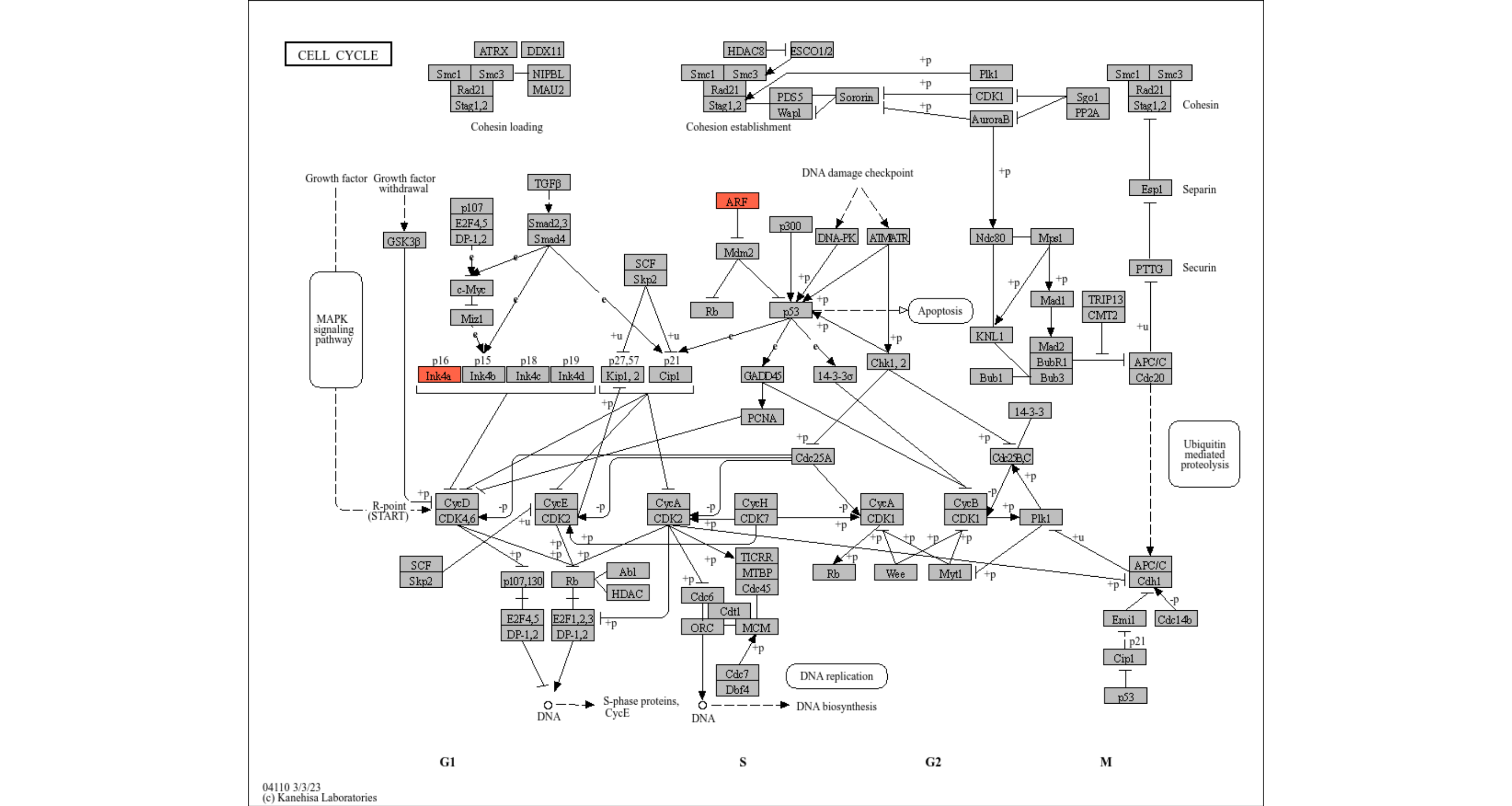
If named numeric vector is provided, continuous scale will be used.
## Highlight genes in the pathway, and overlay raw map.
vecs <- c(-2,2) |> setNames(c("CDKN2A", "CDC45"))
cs <- highlight_entities("hsa04110", vecs) +
scale_fill_viridis(name="LFC")
cs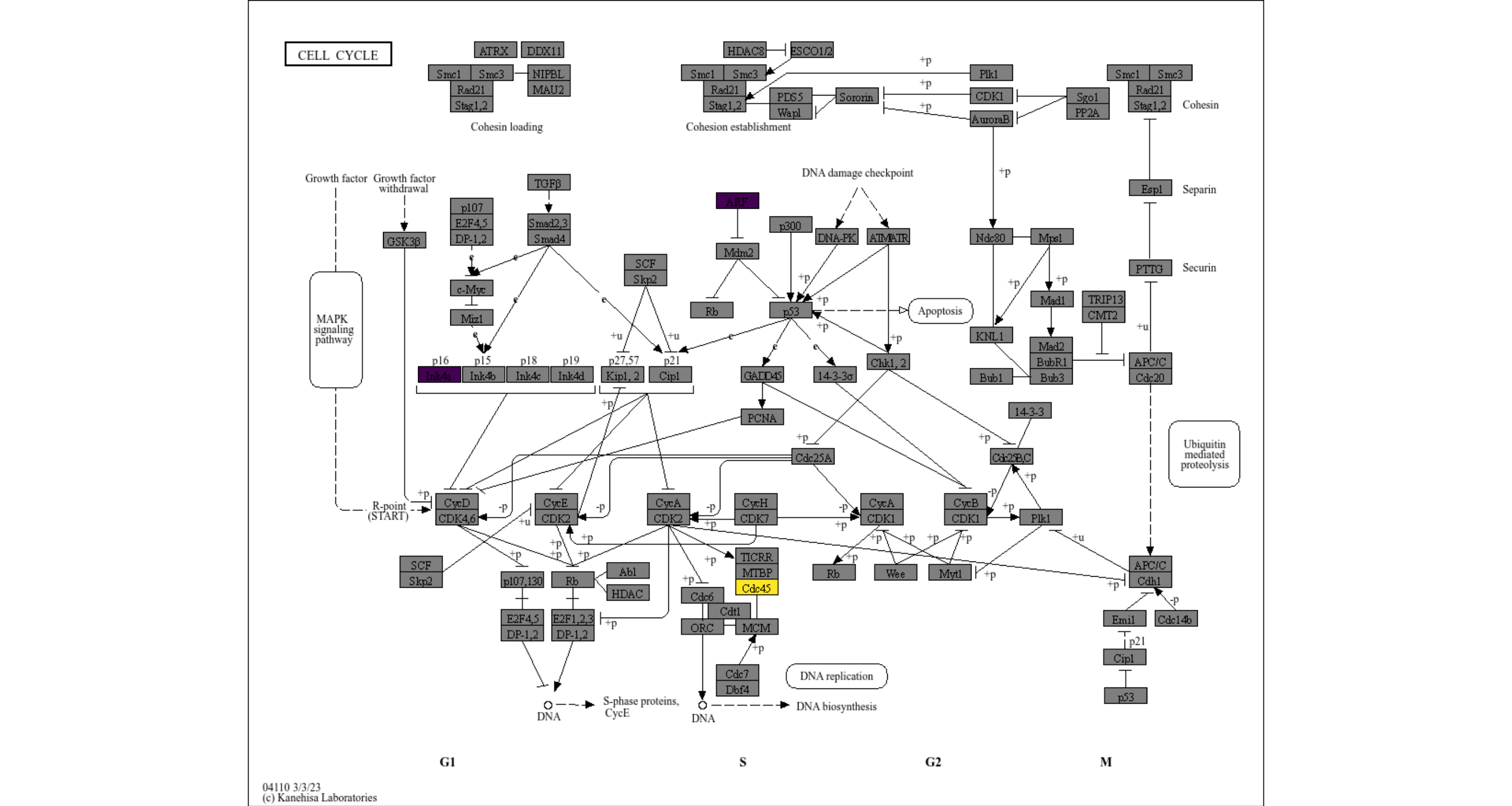
The plot using the original KEGG image can be saved via the ggkeggsave using the original dimension. The function is the wrapper of ggsave.
ggkeggsave(filename="test.png", cs, dpi=300)
knitr::include_graphics("test.png")
1.1 Bugs and errors
If you find bugs, suggestions, or errors such as parsing errors, please kindly report them to Issues, or make a pull request, or report it directly to e-mail.