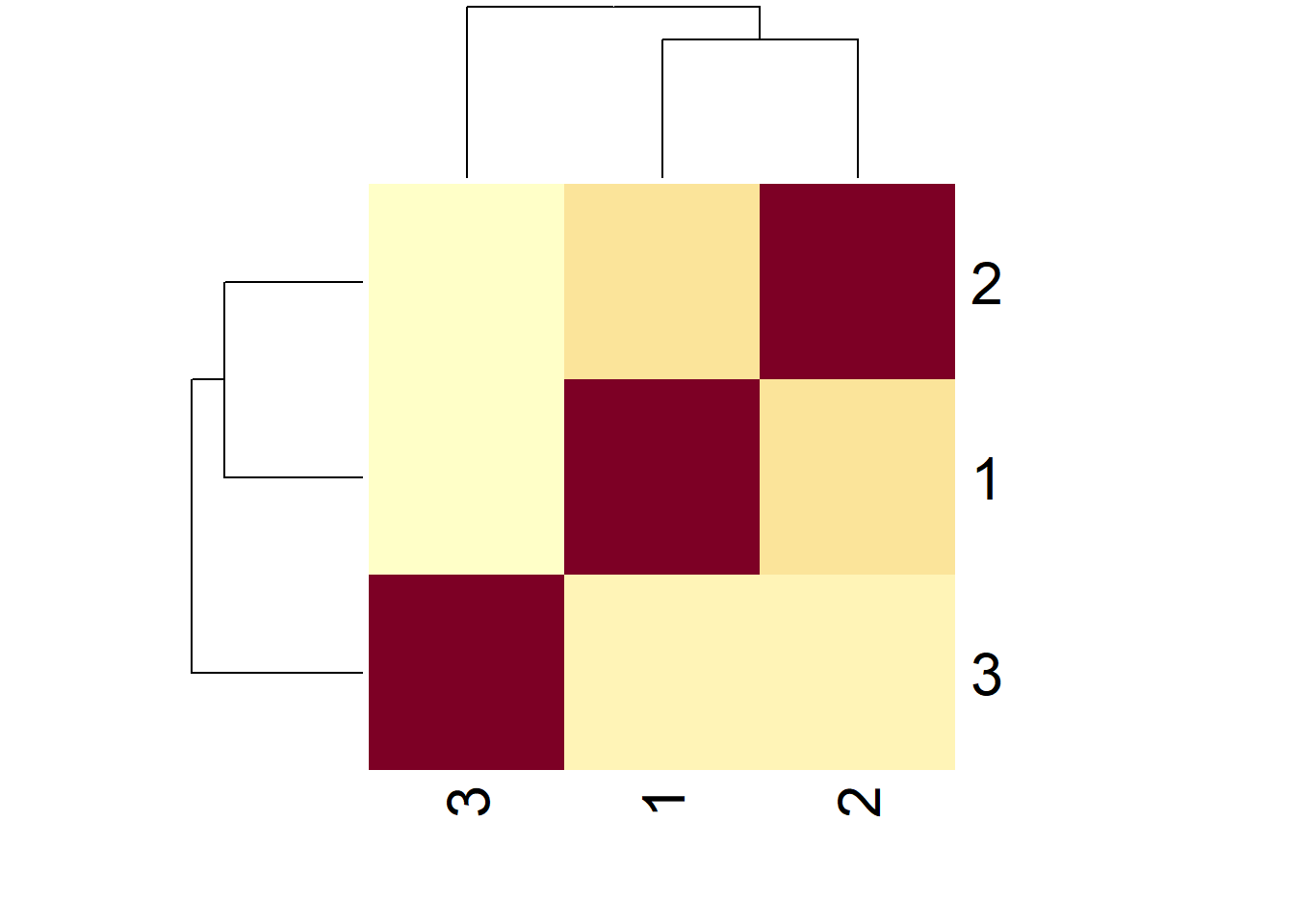7 Quantitative analysis
7.1 Assess the occurrence of the speicific words across gene clusters
library(limma)
library(ggrepel)
query <- "DNA repair"
tab <- getGeneKEGGLinks(species="hsa")
listOfGenes <- list()
for (path in unique(tab$PathwayID)){
listOfGenes[[path]] <- subset(tab, PathwayID==path)$GeneID
}
## Random subset! The results would be different.
frq <- findTerm(query, listOfGenes[sample(length(listOfGenes), 20)],
split=TRUE, calc="mean",
keyType="ENTREZID")
#> Finding query in 20 clusters ...
#> Input genes: 247
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 37
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 97
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 193
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 32
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 137
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 18
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 156
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 108
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 59
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 69
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 3
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 65
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 225
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 30
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 117
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 34
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 50
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 63
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 512
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
plt <- data.frame(t(data.frame(frq, check.names=FALSE)),
check.names=FALSE)
plt$name <- gsub("path:", "", rownames(plt))
p <- ggplot(plt, aes(dna, repair, label = plt[,3])) +
geom_point(color = "red")+
geom_text_repel(bg.color="white")+theme_minimal()+
xlab("dna")+ylab("repair")
p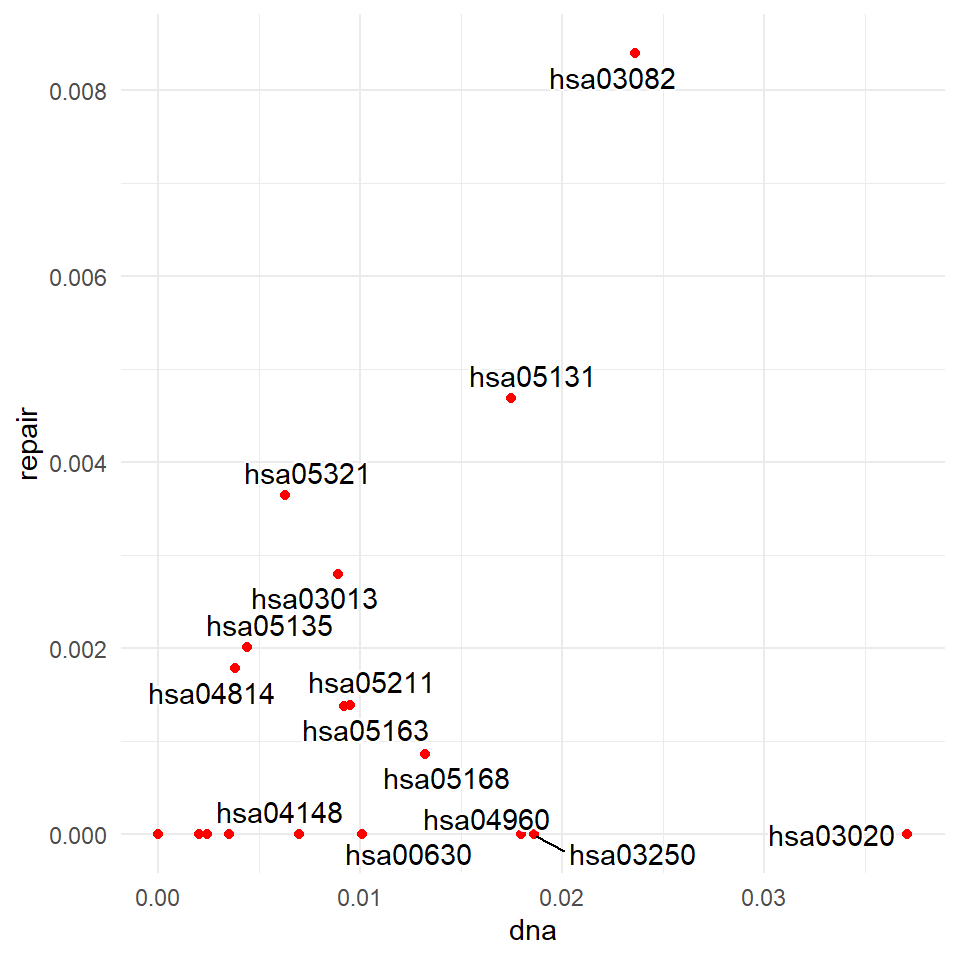
For clustering analysis like WGCNA, making the list and query.
query <- "antiviral response"
load("./blockwiseModule.rda")
mecolors <- bwmod$color
inputList <- names(mecolors)
names(inputList) <- paste0("ME",bwmod$color)
listOfGenes <- split(inputList, names(inputList))
frq <- findTerm(query, listOfGenes,
split=TRUE,calc="highest",
keyType="ENSEMBL")
#> Finding query in 17 clusters ...
#> Input genes: 12526
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 9512
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 5586
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 4595
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 94
#> 'select()' returned 1:1 mapping between keys and
#> columns
#> Converted input genes: 92
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 87
#> 'select()' returned 1:1 mapping between keys and
#> columns
#> Converted input genes: 85
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 86
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 47
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 83
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 77
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 68
#> 'select()' returned 1:1 mapping between keys and
#> columns
#> Converted input genes: 67
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 45
#> 'select()' returned 1:1 mapping between keys and
#> columns
#> Converted input genes: 45
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 36
#> 'select()' returned 1:1 mapping between keys and
#> columns
#> Converted input genes: 9
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 2396
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 2199
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 1066
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 885
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 774
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 626
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 379
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 338
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 233
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 141
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 181
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 168
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 147
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 126
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Input genes: 113
#> 'select()' returned 1:many mapping between keys and
#> columns
#> Converted input genes: 104
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
plt <- data.frame(t(data.frame(frq, check.names=FALSE)),
check.names=FALSE)
plt$name <- row.names(plt)
p <- ggplot(plt, aes(antiviral, response, label = plt[,3])) +
geom_point(color = "blue")+
geom_text_repel(bg.color="white")+theme_minimal()+
xlab("antiviral")+ylab("response")
p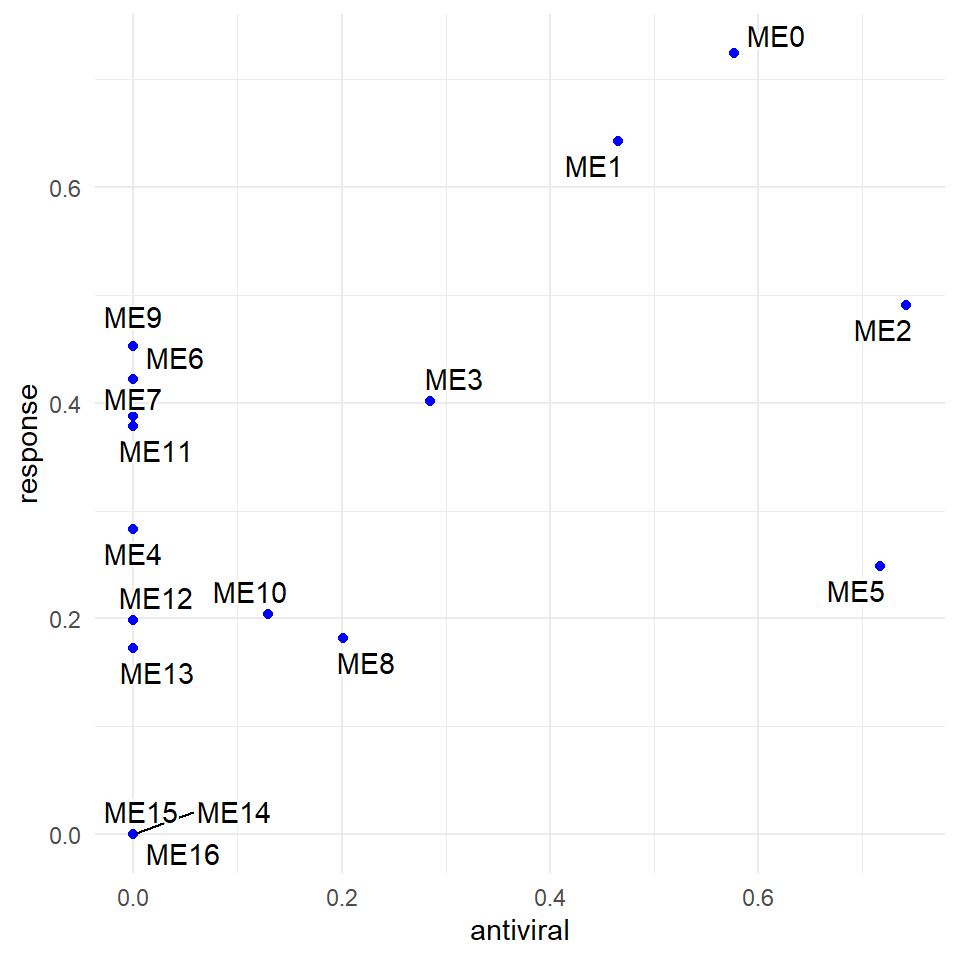
7.2 Recluster the cluster using word information
simExample <- returnSim(returnExample()$color,
keyType="ENSEMBL", argList=list(ora=TRUE))
#> Number of clusters: 3
#> 1
#> Input genes: 12
#> Converted input genes: 7
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Performing ORA
#> Filtered 109 words (ORA)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.6
#> 2
#> Input genes: 13
#> Converted input genes: 13
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Performing ORA
#> Filtered 238 words (ORA)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.4
#> 3
#> Input genes: 7
#> Converted input genes: 7
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Performing ORA
#> Filtered 148 words (ORA)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.5
heatmap(simExample)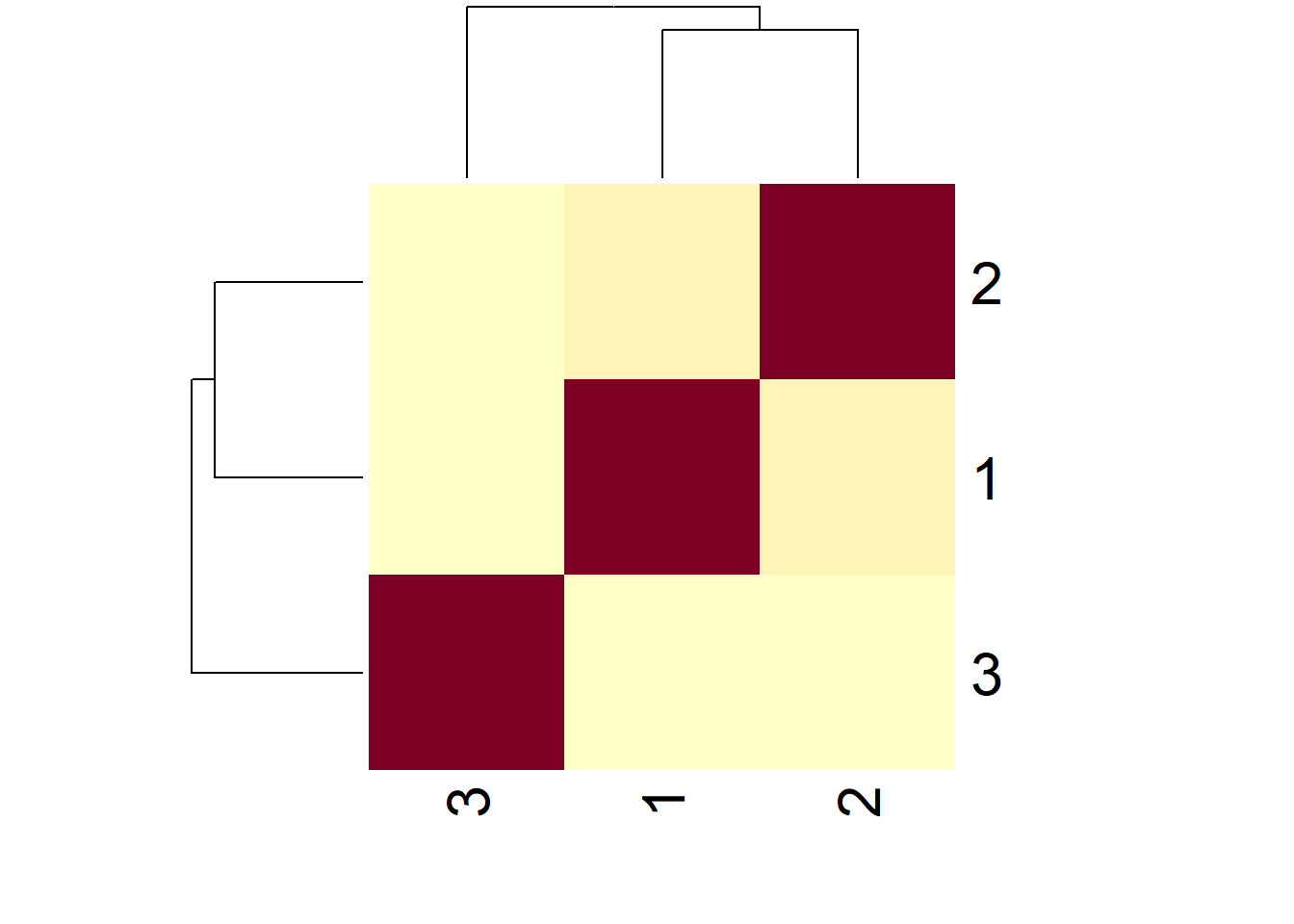
simExample <- returnSim(returnExample()$color,
keyType="ENSEMBL",
argList=list(tfidf=TRUE, takeMax=TRUE))
#> Number of clusters: 3
#> 1
#> Input genes: 12
#> Converted input genes: 7
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.917
#> 2
#> Input genes: 13
#> Converted input genes: 13
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.091
#> 3
#> Input genes: 7
#> Converted input genes: 7
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.216
heatmap(simExample)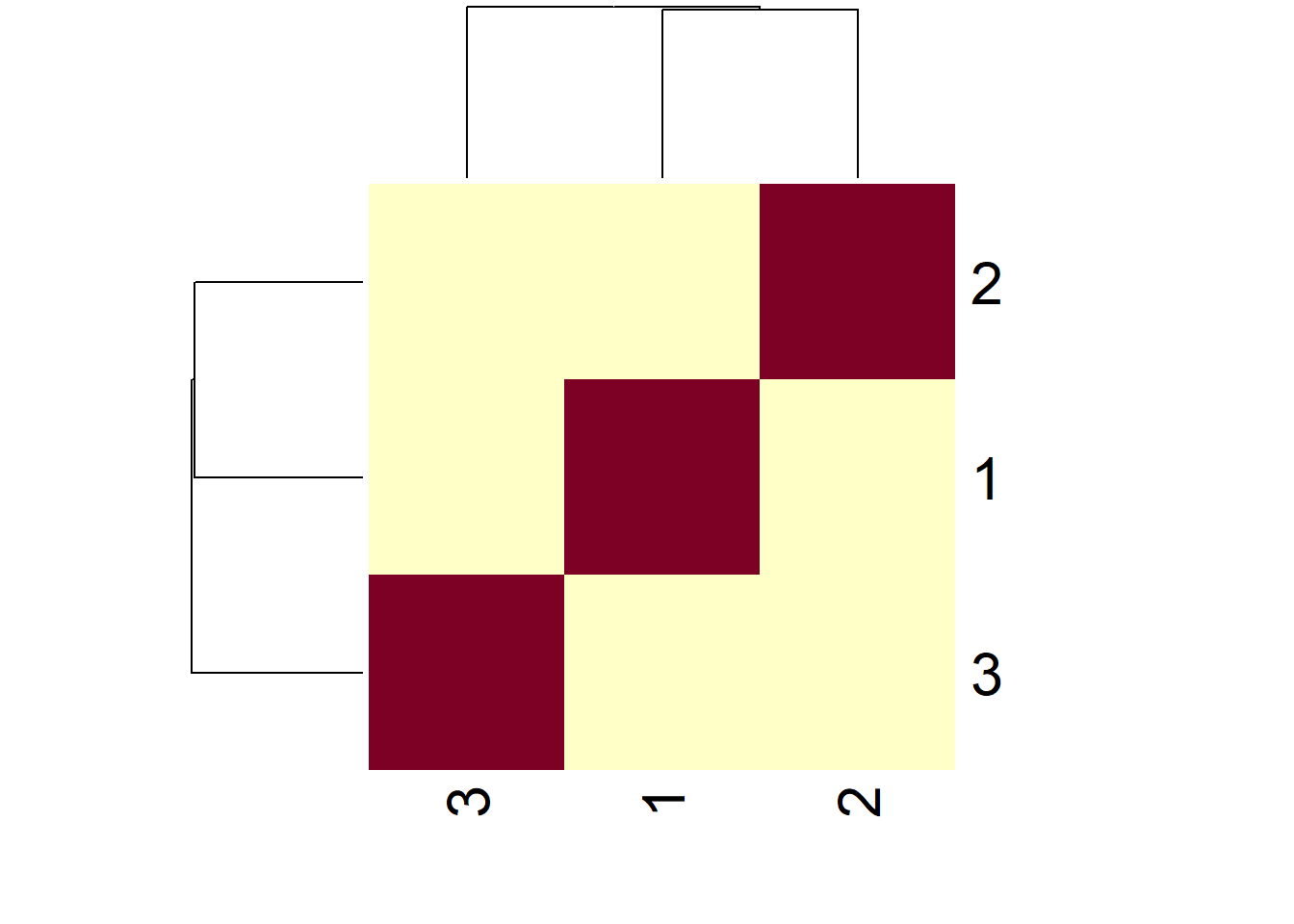
simExample <- returnSim(returnExample()$color,
keyType="ENSEMBL",
argList=list(tfidf=FALSE,
normalize=TRUE,
takeMean=TRUE))
#> Number of clusters: 3
#> 1
#> Input genes: 12
#> Converted input genes: 7
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.6
#> 2
#> Input genes: 13
#> Converted input genes: 13
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.4
#> 3
#> Input genes: 7
#> Converted input genes: 7
#> Filter based on GeneSummary
#> Filtered 77 words (frequency and/or tfidf)
#> Ignoring corThresh, automatically determine the value
#> threshold = 0.7
heatmap(simExample)