Chapter 6 The other options
6.2 Discretization
If passed an option disc=TRUE, the continuous variables are discretized using arules::discretize function (Hahsler et al. 2011). The discretization of the gene expression data is discussed in Gallo et al. (2016). If the same discretization is to be applied on the other data like the training and test dataset, you can pass the training samples to tr option, and if some variables are not intended to be discretized, you should pass the column name to remainCont.
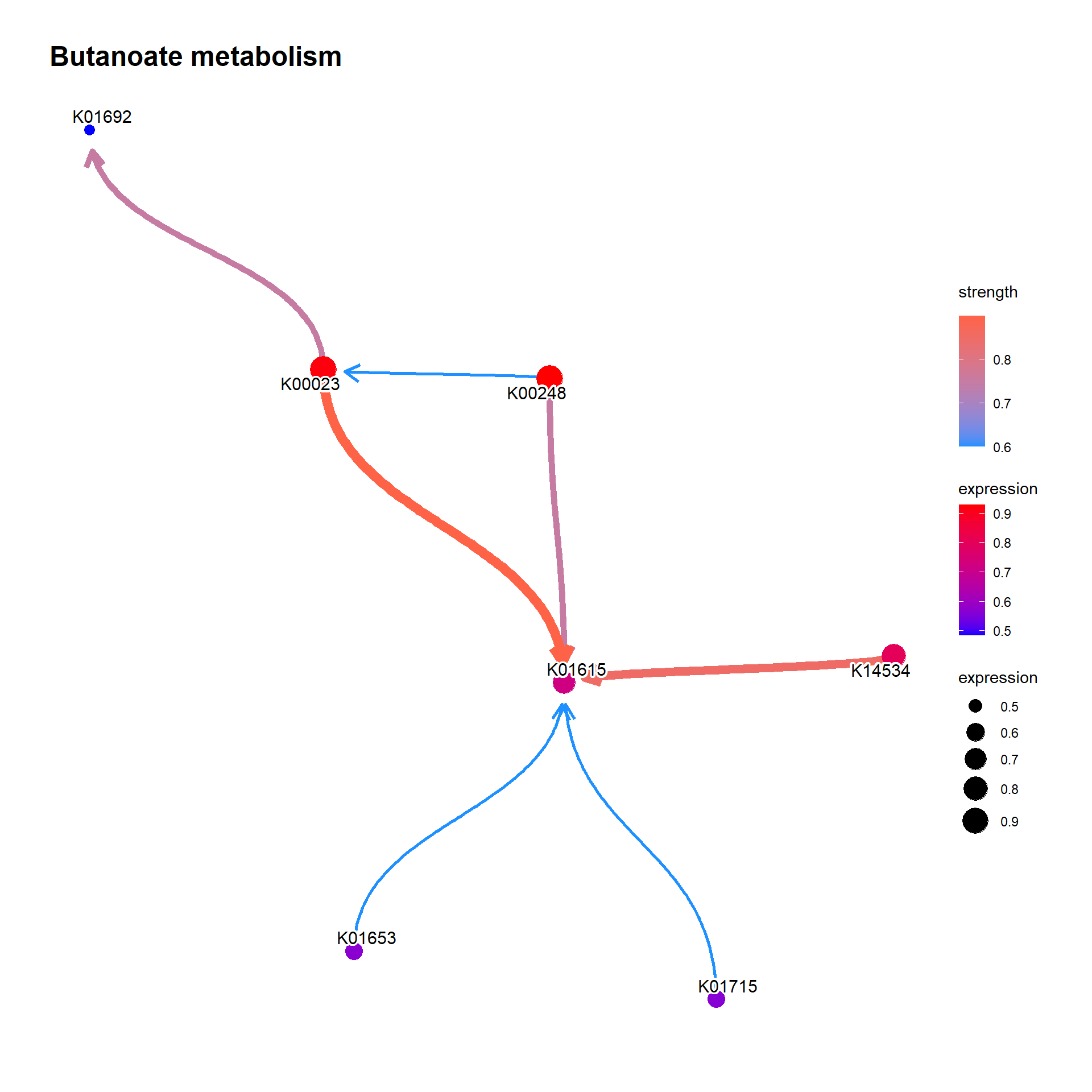
bngeneplot(results = pway, exp = vsted, pathNum = 1, disc=TRUE, layout="sugiyama")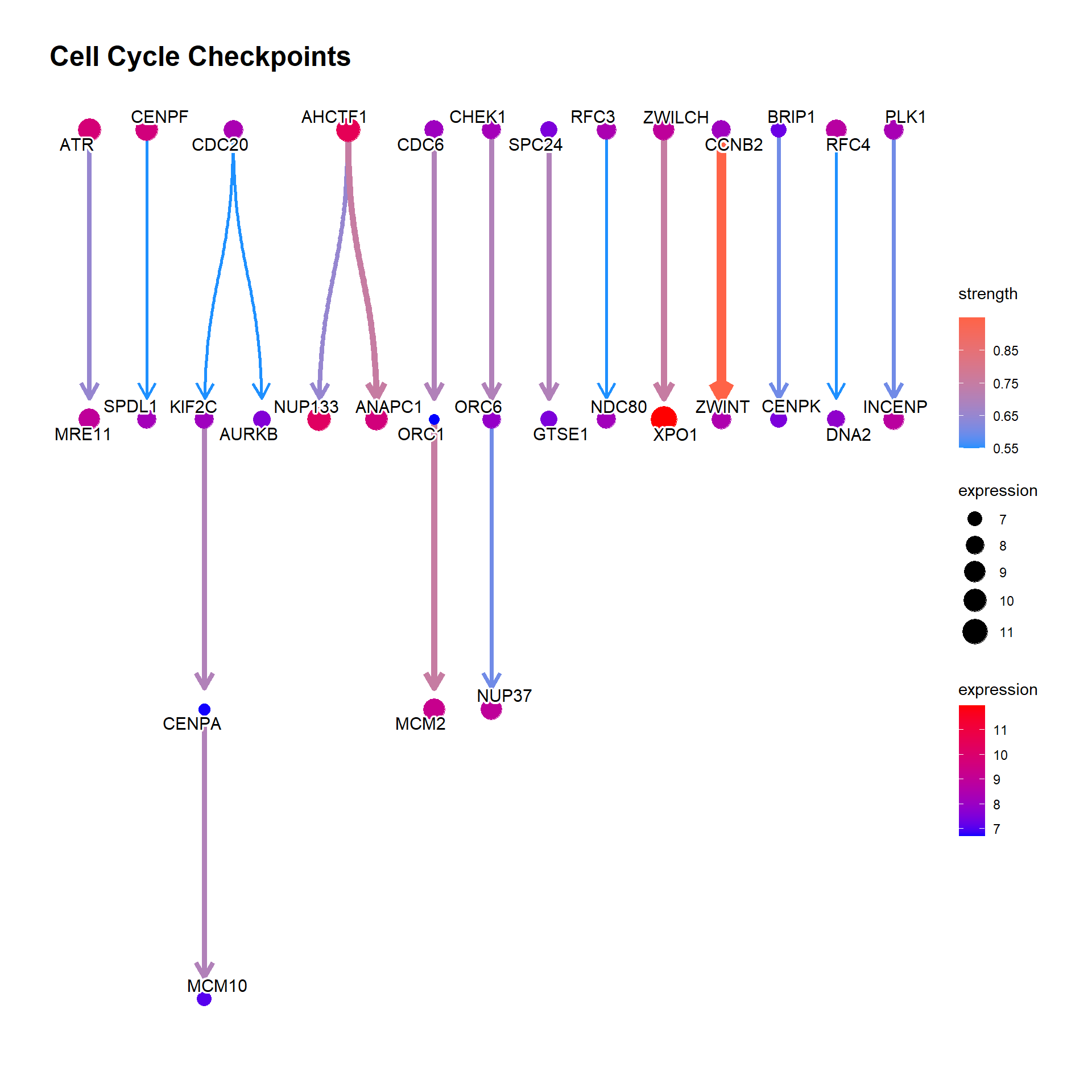
6.3 Custom visualization
6.3.1 The glowing nodes and edges
In addition to the normal plot, custom function of visualization is implemented (bngeneplotCustom and bnpathplotCustom). For example, to effectively visualize the hub genes and edges with high strength by glowing the respective nodes and edges, below is an example using an idea of ggCyberPunk. Additionally, the edge and node colors are fully customizable.
cl = parallel::makeCluster(6)
bngeneplotCustom(results = pway,
exp = vsted,
expSample = incSample,
R=50, cl=cl, layout="nicely", fontFamily="sans",
pathNum = c(4), strType="normal", sizeDep=TRUE, dep=dep,
showDir=FALSE, hub=5, glowEdgeNum=5,
strThresh=0.6, strengthPlot = TRUE)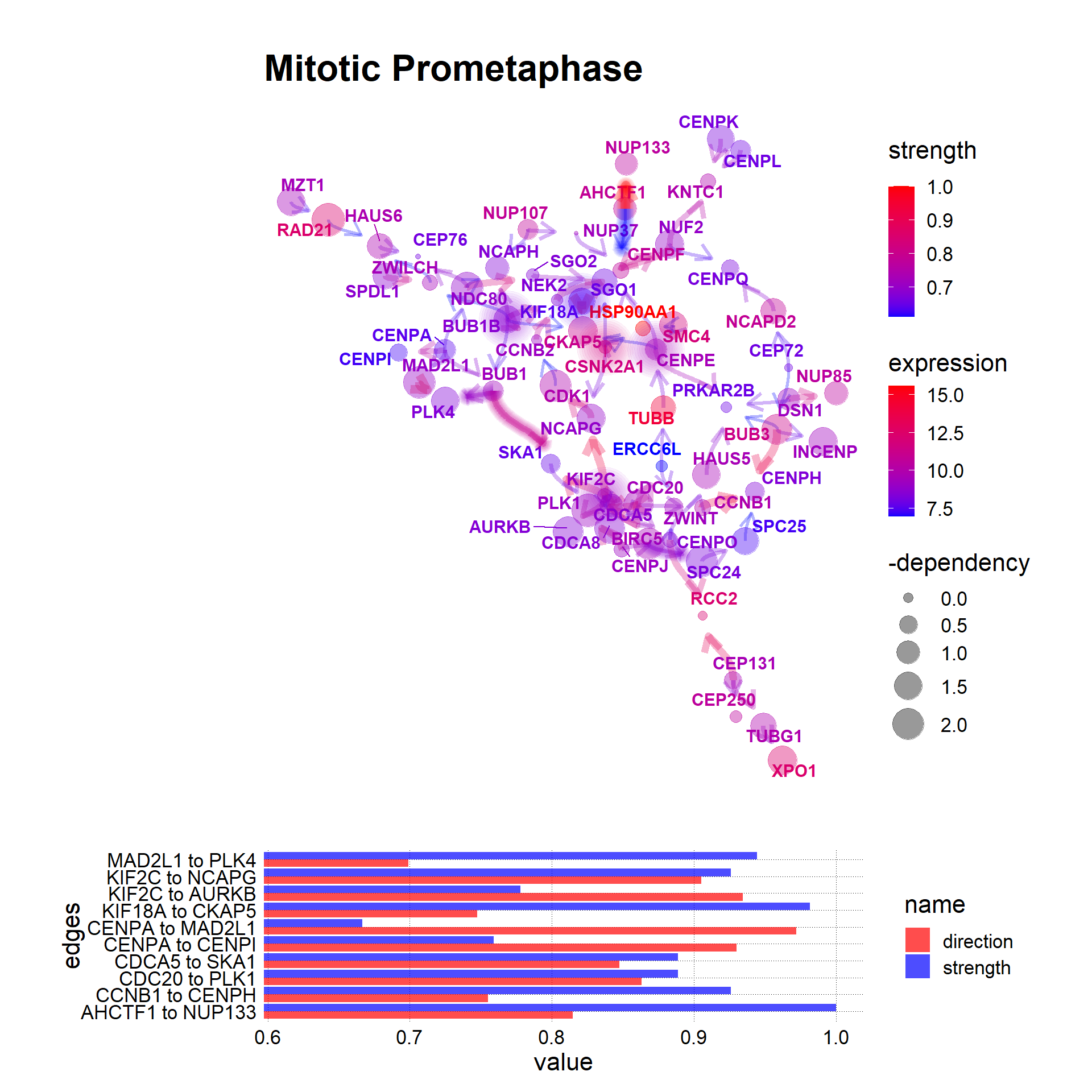
For the demonstrative purpose, using the palettes and fonts of vapoRwave and showtext, the other visualizations are possible. Note that in custom visualization, only the network plot and strength barplot are supported.
## Use alien encounter fonts (http://www.hipsthetic.com/alien-encounters-free-80s-font-family/)
sysfonts::font_add(family="alien",regular="SFAlienEncounters.ttf")
showtext::showtext_auto()
cl = parallel::makeCluster(6)
bngeneplotCustom(results = pway,
exp = vsted,
expSample = incSample,
R=20, cl=cl, fontFamily="alien", labelSize=6,
pathNum = c(15), strType="normal",
showDir=F, hub=5, glowEdgeNum=5, strThresh=0.6,
strengthPlot = T, sizeDep=F, dep=dep, layout="kk",
edgePal=c("#9239F6","#FF4373"),
nodePal=c("#F8B660","#FF0076"),
barLegendKeyCol="#0F0D1A",
textCol="#EE9537", titleCol="#EE9537",
backCol="#0F0D1A",
barAxisCol="#EE9537",
barTextCol="#EE9537",
barPal=c("#9239F6", "#FF4373"),
barPanelGridCol="#FFB967",
barBackCol="#0F0D1A",
titleSize=14
)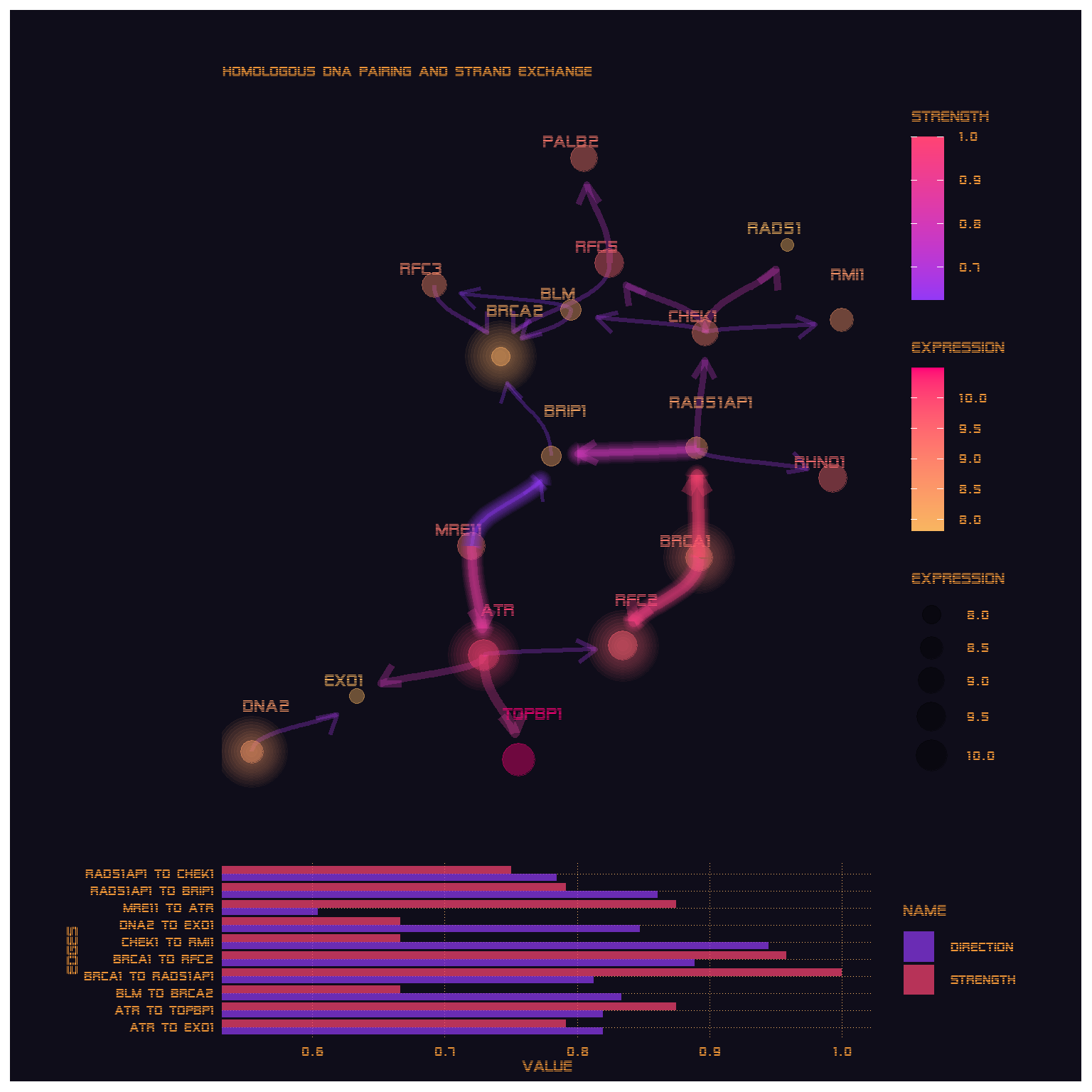
parallel::stopCluster(cl)6.4 Comparing multi scale and standard bootstrapping
cl <- parallel::makeCluster(6)
comparePlot <- bngeneplot(results = pway,
exp = vsted, cl=cl, strType="normal",
pathNum = 15, R = 50, returnNet=T,
shadowText = TRUE)
comparePlotMS <- bngeneplot(results = pway,
exp = vsted, cl=cl, strType="ms",
pathNum = 15, R = 50, returnNet=T,
shadowText = TRUE)
kable(comparePlot$str %>%
filter(direction>0.5) %>%
arrange(desc(strength)) %>%
head())| from | to | strength | direction |
|---|---|---|---|
| TOPBP1 | ATR | 0.9814815 | 0.8657407 |
| BRCA1 | RAD51AP1 | 0.9629630 | 0.5243056 |
| ATR | RFC2 | 0.9444444 | 0.5648148 |
| RFC5 | XRCC3 | 0.9074074 | 0.5160384 |
| CHEK1 | RAD51 | 0.8518519 | 0.8022487 |
| RAD51AP1 | CHEK1 | 0.8333333 | 0.7468585 |
kable(comparePlotMS$str %>%
filter(direction>0.5) %>%
arrange(desc(strength)) %>%
head())| from | to | strength | direction |
|---|---|---|---|
| BRCA1 | RAD51AP1 | 0.9974312 | 0.5885859 |
| RFC2 | ATR | 0.9968008 | 0.6932430 |
| BRIP1 | BRCA2 | 0.9922096 | 0.7125908 |
| DNA2 | BLM | 0.9903698 | 0.5422446 |
| BLM | DNA2 | 0.9903698 | 0.8090673 |
| TOPBP1 | RMI1 | 0.9892198 | 0.5154026 |