Single-cell transcriptomics¶
Provided you completed the 3k PBMCs tutorial of scanpy, we illustrate the depiction of marker gene information upon the KEGG pathway in this example.
[1]:
import numpy as np
import pandas as pd
import scanpy as sc
import matplotlib
import warnings
warnings.filterwarnings('ignore')
matplotlib.rcParams['figure.figsize'] = (6, 6)
[2]:
adata = sc.read_10x_mtx(
'../filtered_gene_bc_matrices/hg19', # the directory with the `.mtx` file
var_names='gene_symbols')
[3]:
adata.var_names_make_unique()
[4]:
sc.pp.filter_cells(adata, min_genes=200)
sc.pp.filter_genes(adata, min_cells=3)
[5]:
adata.var['mt'] = adata.var_names.str.startswith('MT-') # annotate the group of mitochondrial genes as 'mt'
sc.pp.calculate_qc_metrics(adata, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
[6]:
adata = adata[adata.obs.n_genes_by_counts < 2500, :]
adata = adata[adata.obs.pct_counts_mt < 5, :]
[7]:
sc.pp.normalize_total(adata, target_sum=1e4)
[8]:
sc.pp.log1p(adata)
[9]:
sc.pp.highly_variable_genes(adata, min_mean=0.0125, max_mean=3, min_disp=0.5)
[10]:
adata = adata[:, adata.var.highly_variable]
[11]:
sc.pp.regress_out(adata, ['total_counts', 'pct_counts_mt'])
[12]:
sc.pp.scale(adata, max_value=10)
[13]:
sc.tl.pca(adata, svd_solver='arpack')
[14]:
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
[15]:
sc.tl.leiden(adata)
[16]:
sc.tl.rank_genes_groups(adata, 'leiden', method='t-test')
[17]:
## Consider top-50 markers
markers = pd.DataFrame(adata.uns['rank_genes_groups']['names']).head(50)
markers
[17]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
| 0 | LTB | CST3 | HLA-DPB1 | CCL5 | FCER1G | NKG7 | HLA-DPA1 | PF4 |
| 1 | IL32 | TYROBP | CD79A | NKG7 | AIF1 | CTSW | HLA-DPB1 | PPBP |
| 2 | CD2 | S100A8 | HLA-DRB1 | IL32 | LST1 | GZMB | HLA-DRB1 | SDPR |
| 3 | MAL | FCN1 | HLA-DPA1 | CST7 | TYROBP | GNLY | HLA-DMA | GNG11 |
| 4 | AQP3 | LGALS1 | CD79B | GZMA | CST3 | PRF1 | HLA-DQA1 | NRGN |
| 5 | HINT1 | S100A6 | HLA-DQA1 | CTSW | SAT1 | CST7 | HLA-DQB1 | SPARC |
| 6 | GIMAP7 | LGALS2 | MS4A1 | GZMK | COTL1 | GZMA | CST3 | CCL5 |
| 7 | GIMAP5 | AIF1 | HLA-DQB1 | PTPRCAP | IFITM3 | FGFBP2 | FCER1A | RGS18 |
| 8 | LDLRAP1 | GPX1 | CD37 | LYAR | CTSS | FCGR3A | TACR2 | GPX1 |
| 9 | TRABD2A | LST1 | TCL1A | KLRG1 | LGALS1 | SRGN | FSTL1 | TPM4 |
| 10 | ACAP1 | GSTP1 | HLA-DMA | GZMH | FCGR3A | CD247 | SYTL4 | GP9 |
| 11 | FYB | TYMP | LTB | PRF1 | S100A11 | SPON2 | RP11-324I22.4 | HIST1H2AC |
| 12 | TRAF3IP3 | CTSS | LINC00926 | CD2 | HLA-DPA1 | UBB | FAXDC2 | CD9 |
| 13 | ITM2A | FCER1G | HLA-DMB | SAMD3 | TYMP | FCER1G | HLA-DMB | AP001189.4 |
| 14 | PASK | COTL1 | HVCN1 | HOPX | NPC2 | ID2 | HP | TUBB1 |
| 15 | SCGB3A1 | S100A11 | EAF2 | APOBEC3G | CEBPB | CCL4 | RP4-781K5.2 | ITGA2B |
| 16 | TNFRSF4 | SAT1 | IRF8 | TIGIT | HLA-DRB1 | TYROBP | PPT2-EGFL8 | MPP1 |
| 17 | GIMAP4 | CYBA | FCRLA | NCR3 | S100A6 | HOPX | HIST1H2AH | CLU |
| 18 | RP11-18H21.1 | NPC2 | HLA-DOB | GIMAP7 | CYBA | GZMH | GPX1 | TMEM40 |
| 19 | CISH | GRN | PKIG | CCL4 | SRGN | CLIC3 | SAMD14 | CA2 |
| 20 | RP11-291B21.2 | MS4A6A | SMIM14 | CD247 | PYCARD | XCL2 | CLEC10A | ARPC1B |
| 21 | PTPRCAP | LGALS3 | CD72 | ID2 | IFI30 | CCL5 | PROK2 | NCOA4 |
| 22 | GPR183 | PYCARD | P2RX5 | FGFBP2 | HLA-DPB1 | PTPRCAP | PVALB | PTCRA |
| 23 | SELL | TALDO1 | SPIB | ZAP70 | ARPC1B | IGFBP7 | GSTP1 | TREML1 |
| 24 | SRSF5 | FCGRT | PTPRCAP | ARPC5L | ABI3 | AKR1C3 | HGD | FERMT3 |
| 25 | ATP6V0E2 | SRGN | PNOC | C9orf142 | FCN1 | APOBEC3G | ITGA2B | PGRMC1 |
| 26 | IL23A | NCF2 | BLNK | GIMAP4 | APOBEC3A | TTC38 | LINC00957 | SAT1 |
| 27 | ICOS | ARPC1B | KIAA0125 | LITAF | C1orf162 | CYBA | TDRP | LAMTOR1 |
| 28 | NAP1L4 | HLA-DRB1 | SWAP70 | HSP90AA1 | CDA | PRSS23 | LGALS2 | ODC1 |
| 29 | TNFRSF25 | CDA | CD19 | FCRL6 | C5AR1 | ARPC5L | RP11-252A24.3 | ACRBP |
| 30 | SIRPG | IFI30 | IGLL5 | KLRB1 | MAFB | PDIA3 | TGFB1I1 | SEPT5 |
| 31 | AKTIP | C1orf162 | LIMD2 | PRR5 | FGL2 | EFHD2 | GRN | MMD |
| 32 | CD96 | IFITM3 | ADAM28 | GYG1 | TNFRSF1B | S1PR5 | RAB6B | F13A1 |
| 33 | ILF3-AS1 | ALDH2 | SNX3 | JAKMIP1 | BLVRA | LITAF | S100P | SNCA |
| 34 | RIC3 | TNFSF13B | SNX29P2 | STK17A | BLOC1S1 | XCL1 | PBLD | SRGN |
| 35 | PPP1R2 | FPR1 | LAT2 | S1PR5 | TNFSF10 | GPR56 | PDZK1IP1 | LY6G6F |
| 36 | GPR171 | CEBPB | ARHGAP24 | SRGN | LGALS3 | ABI3 | DNAJC27 | CMTM5 |
| 37 | RORA | ASGR1 | FCGR2B | ARL6IP5 | ARRB2 | PLEKHF1 | RP11-390B4.5 | PTGS1 |
| 38 | CCDC66 | LINC00936 | MZB1 | GZMB | NCF2 | HAVCR2 | CCDC122 | GP1BA |
| 39 | RP11-589C21.6 | IGSF6 | PLD4 | PSME1 | TNFSF13B | ZAP70 | ZNF594 | MYL9 |
| 40 | DENND2D | FOLR3 | BTK | TMBIM6 | STX11 | NCR3 | SUOX | RP11-367G6.3 |
| 41 | SSBP1 | CNPY3 | PRKCB | RORA | FCGR2A | PTGDR | ITGB3 | DYNLL1 |
| 42 | SUCLG2 | SERPINB1 | IGJ | GIMAP5 | OAS1 | SAMD3 | SEPT4 | TPM1 |
| 43 | AP3M2 | APOBEC3A | C16orf74 | CD160 | FCGRT | C5orf56 | CYBA | SNX3 |
| 44 | STK17A | PLBD1 | PPP1R14A | SNRPB | CTD-2006K23.1 | PSMB8 | SYP | TALDO1 |
| 45 | MDS2 | BLVRA | RPL22L1 | PTGDR | SYTL4 | CCL3 | CD1C | MARCH2 |
| 46 | CORO1B | S100A12 | FCRL2 | TERF2IP | RGS19 | PSME1 | AP001189.4 | C2orf88 |
| 47 | MAGEH1 | CCL3 | PARP1 | BUB3 | SMCO4 | FCRL6 | PHACTR1 | CLEC1B |
| 48 | SNHG8 | VAMP8 | GUCD1 | ORAI1 | C20orf27 | C9orf142 | DRAXIN | AC147651.3 |
| 49 | CCNG1 | SMCO4 | RP5-887A10.1 | CDC42EP3 | BTK | RAMP1 | COLGALT2 | GAS2L1 |
[18]:
sc.tl.paga(adata)
sc.pl.paga(adata, plot=False) # remove `plot=False` if you want to see the coarse-grained graph
sc.tl.umap(adata, init_pos='paga')
[19]:
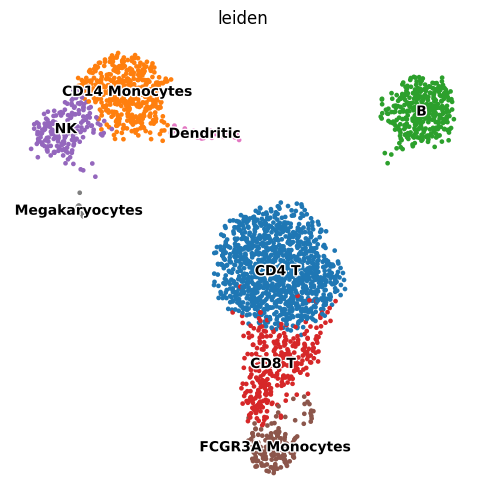
new_cluster_names = [
'CD4 T', 'CD14 Monocytes',
'B', 'CD8 T',
'NK', 'FCGR3A Monocytes',
'Dendritic', 'Megakaryocytes']
# adata.rename_categories('leiden', new_cluster_names)
adata.obs["leiden"] = adata.obs["leiden"].cat.rename_categories(new_cluster_names)
[20]:
sc.pl.umap(adata, color='leiden', legend_loc='on data',
frameon=False, legend_fontsize=10, legend_fontoutline=2)
[21]:
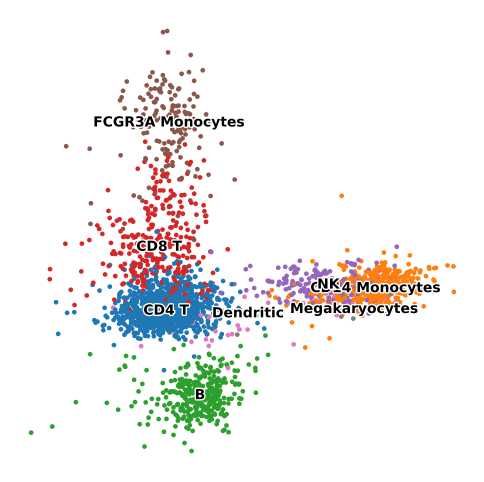
sc.pl.pca(adata, color='leiden', legend_loc='on data', title='', frameon=False, legend_fontoutline=2)
[22]:
import pykegg
from matplotlib import colors as mcolors
from PIL import Image
cols = mcolors.TABLEAU_COLORS
[23]:
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba_array
fig, ax = plt.subplots(1,1)
ax.imshow(to_rgba_array([cols[i] for i in cols.keys()]).reshape(10,1,4)[0:8,:,:])
ax.set_yticks(np.arange(8))
ax.set_yticklabels(new_cluster_names)
plt.tight_layout()
[24]:
import requests_cache
## Cache all the downloaded files
requests_cache.install_cache('pykegg_cache')
[25]:
g = pykegg.KGML_graph(pid="hsa04380")
[26]:
nds = g.get_nodes()
for mark in [0,1,2,3,4,5,6,7]:
nds["in"] = nds["graphics_name"].apply(lambda x:
len(set(markers[str(mark)]) &
set([i.replace(" ","").replace("...","") for i in x.split(",")])))
nds["color"+str(mark)] = [cols[list(cols.keys())[mark]] if i else None for i in nds["in"].tolist()]
[27]:
qc = list()
for id in nds.id:
tmp = nds[nds.id==id].loc[:,[i for i in nds.columns if i.startswith("color")]]
tmpl = tmp.iloc[0,:].tolist()
if len(set(tmpl))==1 and tmpl[0]==None:
qc.append("#ffffff")
else:
qc.append([i for i in tmpl if i is not None])
nds["color"]=qc
[28]:
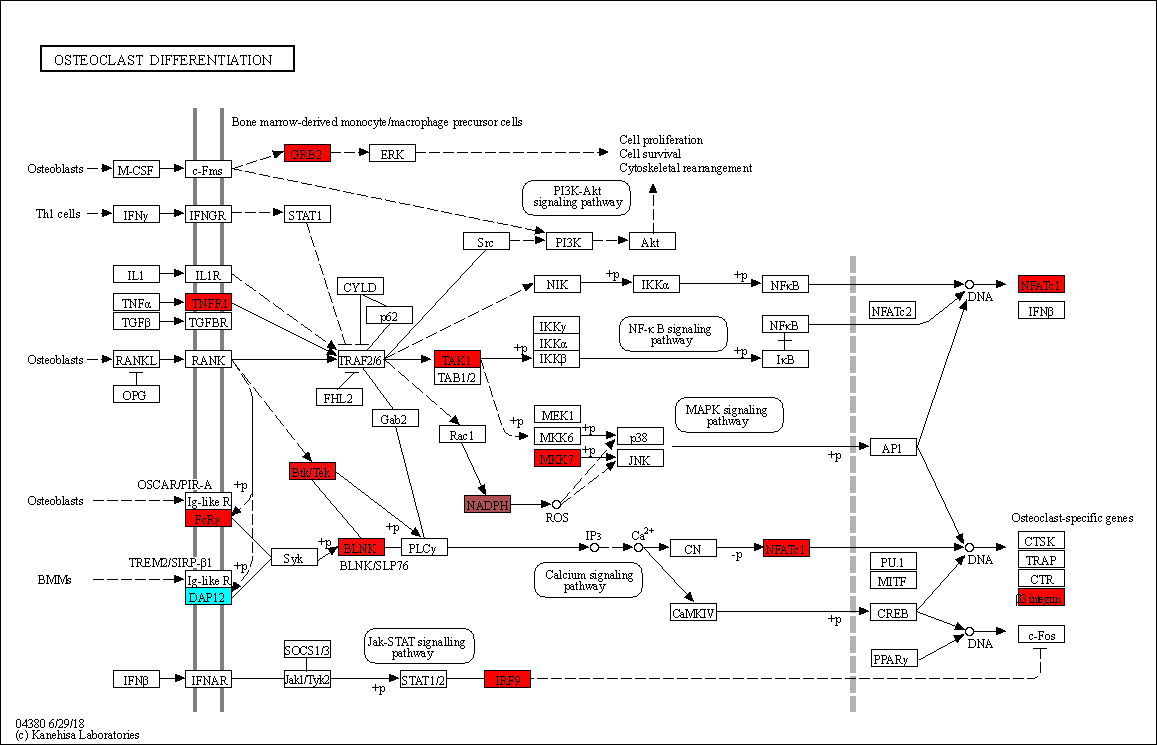
kegg_map = pykegg.overlay_opencv_image(nds, pid="hsa04380")
[29]:
Image.fromarray(kegg_map)
[29]:
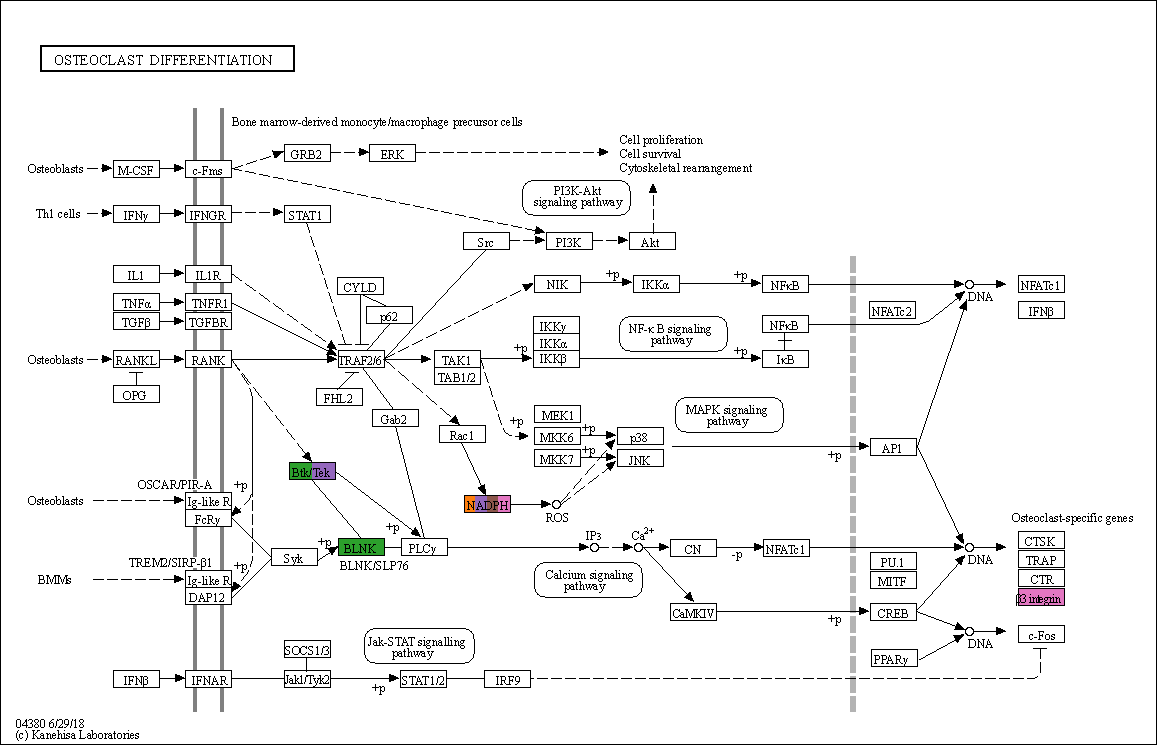
Associate statistical values with the nodes¶
[68]:
## Extract adjusted p-values from the scanpy object
## This time, cluster 4 identified by leiden algorithm
pval_adj = pd.concat([
pd.DataFrame(adata.uns['rank_genes_groups']['names'])["4"],
pd.DataFrame(adata.uns['rank_genes_groups']['pvals_adj'])["4"],
], axis=1)
## Make dict
pval_adj.index = pval_adj.iloc[:,0]
pval_adj_dic = pval_adj.iloc[:,1].apply(lambda x: np.log10(x)).to_dict()
[75]:
## Append new color column based on dict
nds = pykegg.append_colors_continuous_values(nds,
pval_adj_dic,
new_color_column="pval_adj_4",
two_slope=False,
colors=["cyan","red"])
[78]:
img_arr = pykegg.overlay_opencv_image(nds, pid="hsa04380", fill_color="pval_adj_4")
[79]:
## Visualize
Image.fromarray(img_arr)
[79]:
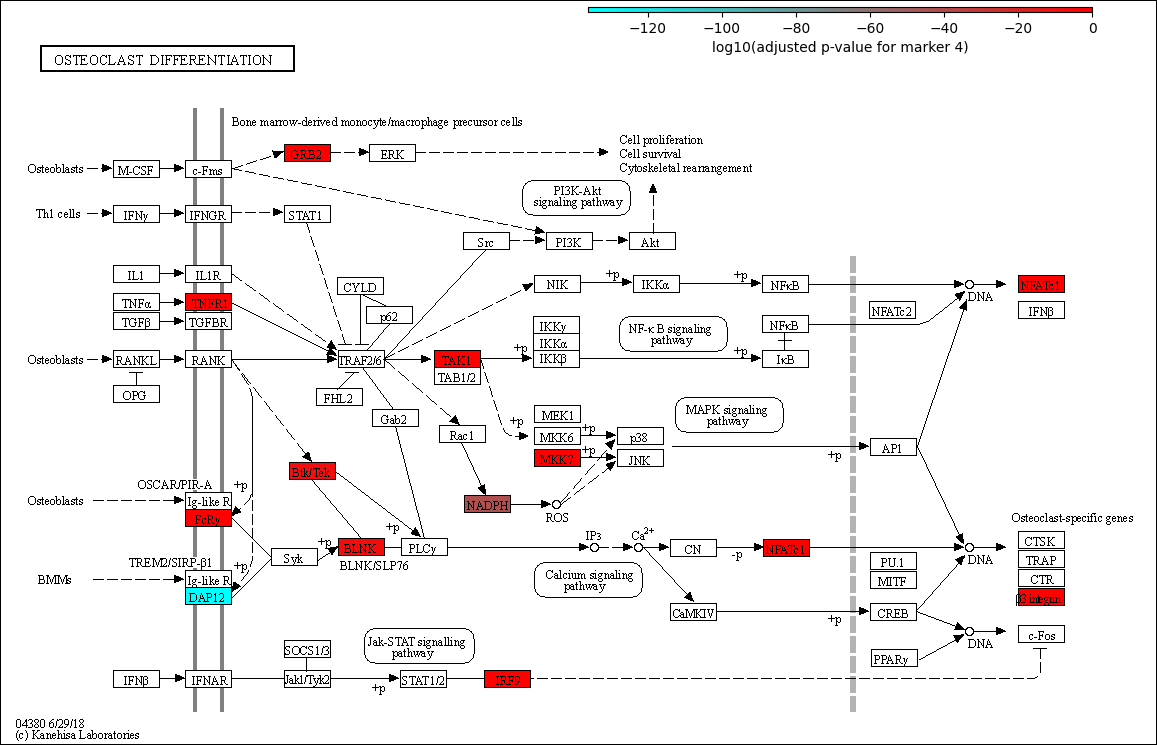
Legends¶
The legend can be added through the append_legend function. Same specifications as when specifying colors are required for the min_value, max_value, and center_value. The position can be selected from four options: “topright,” “topleft,” “bottomright,” and “bottomleft.”
[62]:
max([pval_adj_dic[i] for i in pval_adj_dic.keys()])
[62]:
-0.0006253720313310442
[80]:
Image.fromarray(pykegg.append_legend(img_arr,
min_value=min([pval_adj_dic[i] for i in pval_adj_dic.keys()]),
max_value=0, two_slope=False, colors=["cyan","red"],
width=6.5, label="log10(adjusted p-value for marker 4)"))
[80]:
[ ]: